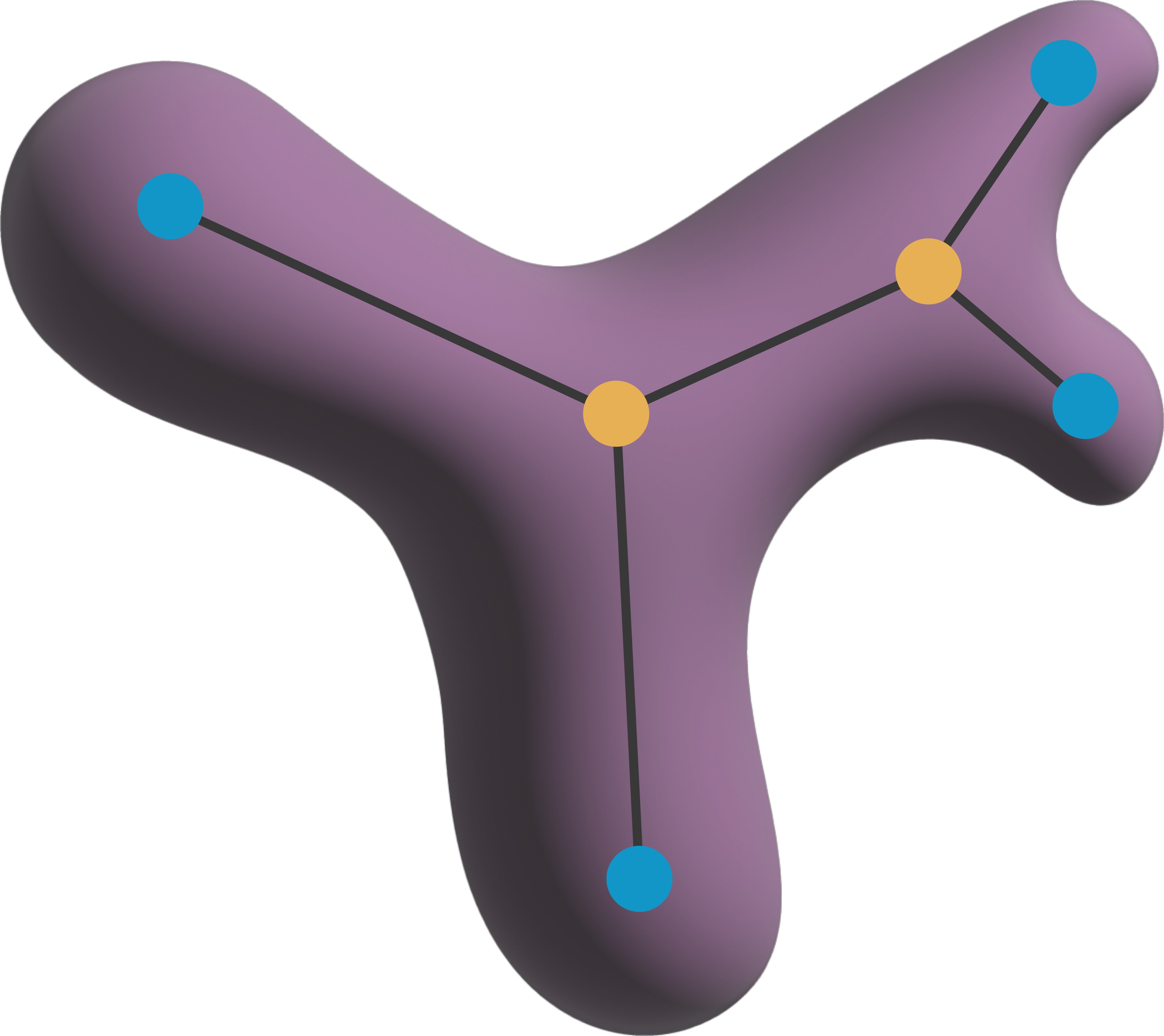
Topological skeleton analysis in 3D#
# import 3d binary blobs
import numpy as np
import napari_toska as nts
from skimage import data, measure
import napari
import networkx as nx
viewer = napari.Viewer(ndisplay=3)
Assistant skips harvesting pyclesperanto as it's not installed.
image = data.binary_blobs(length=64, n_dim=3, rng=0, blob_size_fraction=0.3)
labels = measure.label(image)
skeleton_labeled = nts.generate_labeled_skeletonization(labels)
skeleton_parsed = nts.parse_all_skeletons(skeleton_labeled, neighborhood='n26')
viewer.add_labels(labels, name='blobs')
viewer.add_labels(skeleton_labeled, name='skeleton')
viewer.add_labels(skeleton_parsed, name='parsed skeleton')
<Labels layer 'parsed skeleton' at 0x20f8a8c6a30>
skeleton_parsed_single = skeleton_parsed * (skeleton_labeled == 2)
viewer.add_labels(skeleton_parsed_single, name='parsed_skeleton_single')
<Labels layer 'parsed_skeleton_single' at 0x20f986a7fd0>
# create an adjacency matrix for the skeleton
adjacency_matrix = nts.create_adjacency_matrix(skeleton_parsed_single,
neighborhood='n26')
graph = nts.convert_adjacency_matrix_to_graph(adjacency_matrix)
features_single = nts.analyze_single_skeleton(skeleton_parsed_single, neighborhood='n26')
features_single
| n_endpoints | n_branch_points | n_nodes | n_branches | n_cycle_basis | n_possible_undirected_cycles | |
|---|---|---|---|---|---|---|
| 0 | 10 | 8 | 18 | 18 | 1 | 1 |
labeled_branches_single = nts.label_branches(skeleton_parsed_single, skeleton_labeled, neighborhood='n26')
viewer.add_labels(labeled_branches_single, name='labeled_branches')
<Labels layer 'labeled_branches' at 0x20f9890d1f0>
spine_image = nts.create_spine_image(adjacency_matrix=adjacency_matrix, labeled_branches=labeled_branches_single)
viewer.add_labels(spine_image, name='spine_image')
<Labels layer 'spine_image' at 0x20f986a7d30>
nts.calculate_branch_lengths(labeled_branches_single)
| label | branch_length | |
|---|---|---|
| 0 | 1 | 17.899495 |
| 1 | 2 | 10.242641 |
| 2 | 3 | 3.414214 |
| 3 | 4 | 4.560478 |
| 4 | 5 | 2.828427 |
| 5 | 6 | 20.120956 |
| 6 | 7 | 27.388905 |
| 7 | 8 | 91.698485 |
| 8 | 9 | 13.242641 |
| 9 | 10 | 33.116827 |
| 10 | 11 | 10.974691 |
| 11 | 12 | 1.732051 |
| 12 | 13 | 5.656854 |
| 13 | 14 | 19.999271 |
| 14 | 15 | 20.071068 |
| 15 | 16 | 4.242641 |
| 16 | 17 | 13.242641 |
| 17 | 18 | 3.414214 |
# analyze all skeletons
features = nts.analyze_skeletons(
labeled_skeletons=skeleton_labeled,
parsed_skeletons=skeleton_parsed,
neighborhood='n26')
features
| skeleton_id | n_endpoints | n_branch_points | n_nodes | n_branches | n_cycle_basis | n_possible_undirected_cycles | label | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| 0 | 2 | 10 | 8 | 18 | 18 | 1 | 1 | 2 |