from napari.layers import Labels
import napari_toska as nts
import networkx as nx
import numpy as np
import pandas as pd
[docs]
class ToskaSkeleton(Labels):
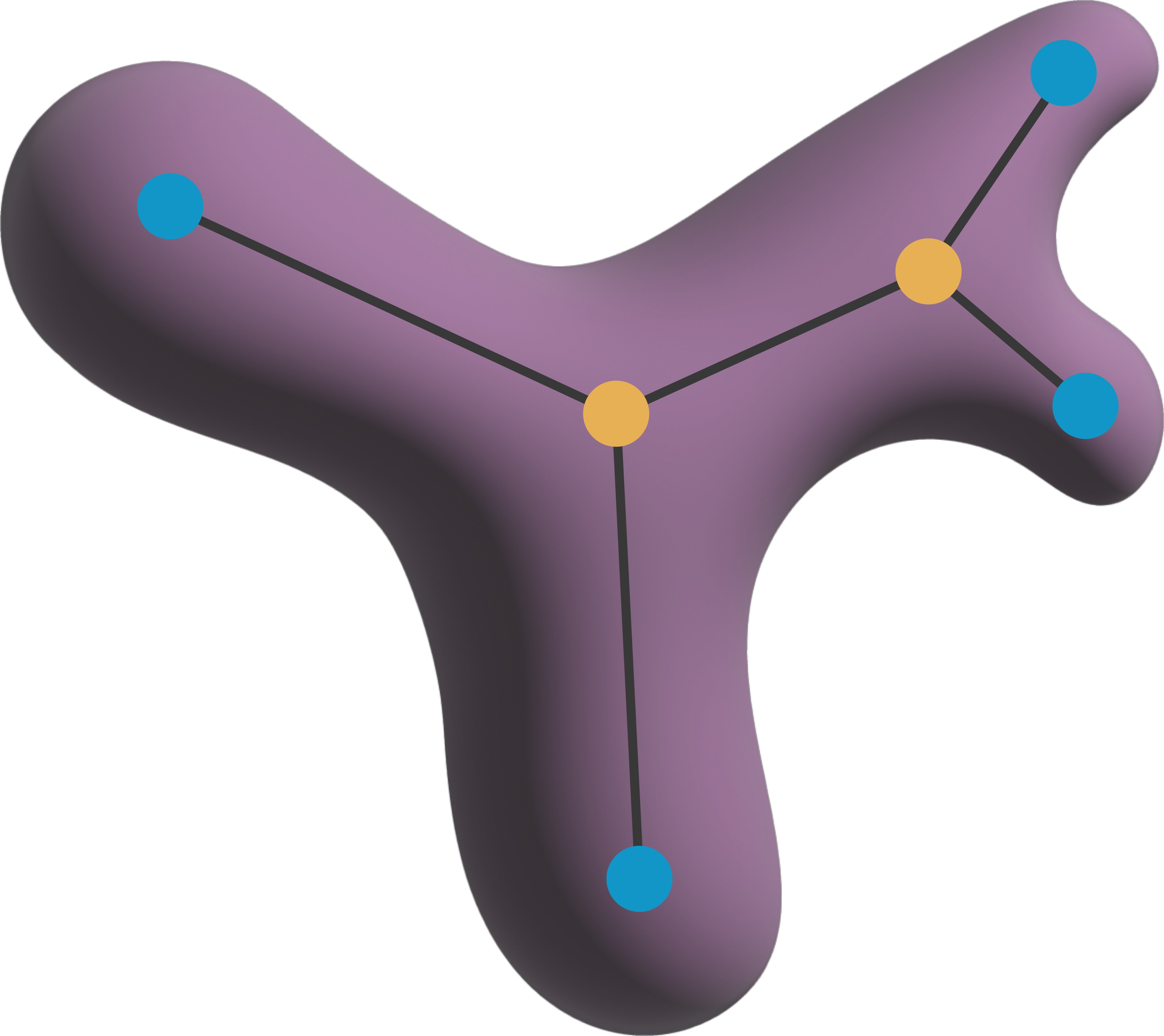
"""
A class to represent a skeleton image in napari.
Parameters:
-----------
labels_data : napari.types.LabelsData
The data to be processed - has to be a napari Label image.
neighborhood : str
The neighborhood connectivity of the skeleton. Can be "n4",
"n6", "n8", "n18", or "n26". "n4" and "n8" apply only to 2D images.
"n6", "n18", and "n26" apply to 3D images.
**kwargs
Additional keyword arguments to pass to the Labels layer.
Attributes:
-----------
neighborhood : str
The neighborhood connectivity of the skeleton.
graph : nx.Graph
A networkx graph representing the skeleton. The graph is built
from the skeleton data and is stored in `layer.metadata` for easy access.
features : pd.DataFrame
A DataFrame containing features of the individual nodes and edges of the skeleton
graph but also of the skeleton as a whole.
Methods:
--------
analyze()
Analyze the skeleton data and build a networkx graph from it.
create_feature_map(feature: str) -> napari.types.ImageData
Create a feature map from the skeleton data. Any column
in the features DataFrame can be used as a feature.
Examples:
---------
>>> import napari
>>> import napari_toska as nts
>>> from skimage.data import binary_blobs
>>> from skimage.measure import label
>>>
>>> # create a binary image
>>> labels = label(binary_blobs(rng=0))
>>>
>>> # Build the Skeleton object
>>> Skeleton = nts.ToskaSkeleton(labels, neighborhood='n8')
>>> Skeleton.analyze()
"""
def __init__(self, labels_data: "napari.types.LabelsData", neighborhood: str, **kwargs):
super().__init__(np.asarray(labels_data), **kwargs)
self._neighborhood = neighborhood
self.graph = nx.Graph()
def analyze(self):
"""
Analyze the skeleton data and build a networkx graph from it.
Returns:
--------
None
"""
self._parse_skeleton()
self._build_nx_graph()
self._measure_branch_length()
self._detect_spines()
self._graph_summary()
# drop index column
if 'index' in self.features.columns:
self.features = self.features.drop(columns='index')
@property
def neighborhood(self):
return self._neighborhood
@property
def graph(self) -> nx.Graph:
return self.metadata.get('graph')
@graph.setter
def graph(self, graph: nx.Graph):
self.metadata['graph'] = graph
def create_feature_map(self, feature: str) -> "napari.types.ImageData":
feature_map = np.zeros(self.data.shape, dtype=int)
for _, row in self.features.iterrows():
feature_map[self.data == row['label']] = row[feature]
return feature_map
def _parse_skeleton(self) -> None:
from skimage import measure
labelled_skeletons = nts.generate_labeled_skeletonization(self.data).astype(int)
parsed_skeletons = nts.parse_all_skeletons(labelled_skeletons, neighborhood=self._neighborhood)
end_points = measure.label(parsed_skeletons == 1)
branches = measure.label(parsed_skeletons == 2)
branch_points = measure.label(parsed_skeletons == 3)
# create a labels image with unique labels for each object
n_endpoints = end_points.max()
n_branches = branches.max()
branches[branches != 0] += n_endpoints
branch_points[branch_points != 0] += n_endpoints + n_branches
unique_labels = (end_points + branches + branch_points).astype(int)
# add object types to features (branch/end/chain)
object_types = np.ones((unique_labels.max()), dtype=int)
object_types[:n_endpoints] = 1 # end points
object_types[n_endpoints:n_endpoints + n_branches] = 2 # branches
object_types[n_endpoints + n_branches:] = 3 # branch points
self.data = unique_labels
self.features = pd.DataFrame({
'label': np.arange(1, unique_labels.max() + 1).astype(int),
'object_type': object_types,
})
# add skeleton ID to features as a categorical column
for i, row in self.features.iterrows():
self.features.loc[i, 'skeleton_id'] = labelled_skeletons[self.data == row['label']][0]
# make column type categorical
self.features['skeleton_id'] = self.features['skeleton_id'].astype('category')
self.features['object_type'] = self.features['object_type'].astype('category')
self.features['label'] = self.features['label'].astype('category')
return
def _build_nx_graph(self):
import tqdm
# add all branch points and end points to Graph
for _, row in self.features[self.features['object_type'] != 2].iterrows():
self.graph.add_node(row['label'],
object_type=row['object_type'],
label=row['label'])
df_branches = self.features[self.features['object_type'] == 2]
# iterate over branches and find neighboring branch points or end points
for i, row in tqdm.tqdm(df_branches.iterrows(), desc='Building Graph', total=len(df_branches)):
connecting_labels = self._find_neighboring_labels(row['label'])
# a branch should connect to exactly two other objects
if len(connecting_labels) != 2:
self.features.iloc[i, self.features.columns.get_loc('object_type')] = 1
print('detected malformatted label: {}'.format(int(row['label'])),
'changed type from 2 (branch) -> 1 (end point)')
continue
self.graph.add_edge(connecting_labels[0], connecting_labels[1],
label=row['label'])
# check if there are any isolated nodes of type 1 (end points)
isolated_nodes = np.array([node for node in self.graph.nodes if self.graph.degree(node) == 0], dtype=int)
if len(isolated_nodes) > 0:
print('Found isolated nodes: ', isolated_nodes)
# check for neighborhood around isolated nodes
for node in isolated_nodes:
connecting_labels = self._find_neighboring_labels(node)
if len(connecting_labels) == 2:
self.graph.add_edge(connecting_labels[0], connecting_labels[1],
label=None)
else:
print('Could not connect isolated node: ', node)
def _measure_branch_length(self):
# create a label images with only branch labels, mute all the others
LUT = np.asarray([0] + list(self.features['label']))
object_type = np.asarray([0] + list(self.features['object_type']))
# set entries to zero where the object type is 1 or 3
LUT[object_type == 1] = 0
LUT[object_type == 3] = 0
branch_label_image = LUT[self.data]
# measure the length of each branch
branch_lengths = nts.calculate_branch_lengths(branch_label_image)
# merge into features
self.features = pd.merge(self.features, branch_lengths, on='label', how='left')
self.features['branch_length'] = self.features['branch_length'].fillna(0)
# update graph edge weights with branch lengths
for u, v, data in self.graph.edges(data=True):
data['branch_length'] = self.features[self.features['label'] == data['label']]['branch_length'].values[0]
def _find_neighboring_labels(self, query_label: int):
from skimage import morphology
branch = self.data == query_label
branch_point_coordinates = np.asarray(np.where(branch)).T
# get bounding box around branch
min_coords = branch_point_coordinates.min(axis=0) - 1
max_coords = branch_point_coordinates.max(axis=0) + 2
# check if min/max values exceed array dimensions
min_coords[min_coords < 0] = 0
for i in range(len(max_coords)):
if max_coords[i] > branch.shape[i]:
max_coords[i] = branch.shape[i]
# Create a tuple of slices for each dimension
slices = tuple(
slice(min_coord, max_coord) for min_coord, max_coord in zip(min_coords, max_coords)
)
# Crop the image data using the slices,
# then expand the branch points to overlap with the neighboring objects
cropped_branch = branch[slices]
cropped_branch = morphology.binary_dilation(cropped_branch, footprint=np.ones((3, 3)))
cropped_data = self.data[slices]
touching_labels = np.logical_xor(cropped_branch, cropped_data == query_label) * cropped_data
connecting_labels = np.unique(touching_labels)
connecting_labels = connecting_labels[connecting_labels != 0]
return connecting_labels
def _detect_spines(self):
"""
Detect spines in the skeleton graph.
The spine is defined as the longest path between two degree 1 nodes in the skeleton graph.
Returns:
--------
None
"""
import tqdm
graph = self.graph.copy()
self.features['spine'] = 0
# split in connected components
connected_components = list(nx.connected_components(graph))
# find degree 1 nodes in connected components
for idx, component in tqdm.tqdm(enumerate(connected_components), total=len(connected_components), desc='Finding spines'):
spine_nodes = []
for node in component:
if graph.degree(node) == 1:
spine_nodes.append(int(node))
if len(spine_nodes) < 2:
continue
# measure distances between every pair of spine nodes
spine_distances = []
for i, spine_node in enumerate(spine_nodes):
for j in range(i+1, len(spine_nodes)):
path = [int(i) for i in nx.shortest_path(graph, source=spine_node, target=spine_nodes[j])]
# measure distance as per the edge attrbitue 'branch_length'
distance = 0
edge_labels = []
for k in range(len(path) - 1):
distance += graph[path[k]][path[k+1]]['branch_length']
edge_labels.append(graph[path[k]][path[k+1]]['label'])
# for k in range(len(path) - 1):
# distance += graph[path[k]][path[k+1]]['branch_length']
spine_distances.append({
'spine_1': spine_node,
'spine_2': spine_nodes[j],
'distance': distance,
'edge_labels': [int(i) for i in edge_labels]
})
# sort by distance
spine_distances = pd.DataFrame(spine_distances)
spine_distances = spine_distances.sort_values(by='distance')
# set spine attribute to 1 for nodes and edges in shortest path
longest_shortest_path = spine_distances.iloc[-1]
#graph.edges[shortest_path['spine_1'], shortest_path['spine_2']]['spine'] = 1
for label in longest_shortest_path['edge_labels']:
self.features.loc[self.features['label'] == label, 'spine'] = 1
# make spine column categorical
self.features['spine'] = self.features['spine'].astype('category')
def _graph_summary(self):
"""
Calculate summary features of the skeleton graph.
The following features are calculated:
- n_branches:
The number of branches in the skeleton.
- n_endpoints:
The number of endpoints in the skeleton.
- n_branch_points:
The number of branch points in the skeleton.
- n_nodes:
The number of nodes in the skeleton, which is the sum of branch points and endpoints.
- n_cycle_basis:
The number of cycle basis in the skeleton. A cycle basis is a set of cycles that
can be used to generate all other cycles in a graph.
- n_possible_undirected_cycles:
The number of possible undirected cycles in the skeleton. An undirected cycle is a
path that starts and ends at the same node and visits each node only
once (except for the starting node).
Returns:
--------
None
"""
import tqdm
# get subgraphs
connected_components = [self.graph.subgraph(c).copy() for c in nx.connected_components(self.graph)]
self.features['n_branches'] = 0
self.features['n_endpoints'] = 0
self.features['n_branch_points'] = 0
self.features['n_nodes'] = 0
for component in tqdm.tqdm(connected_components, desc='Calculating summary features', total=len(connected_components)):
n_branches = len(component.edges)
n_endpoints = len([node for node in component.nodes if component.degree(node) == 1])
n_branchpoints = len([node for node in component.nodes if component.degree(node) > 2])
# cycle basis
directed_graph = component.to_directed()
possible_directed_cycles = list(nx.simple_cycles(directed_graph))
n_cycle_basis = len(nx.cycle_basis(component))
n_possible_undirected_cycles = len(
[x for x in possible_directed_cycles if len(x) > 2])//2
# add to features
# get label property of all edges and nodes
edge_labels = [component[u][v]['label'] for u, v in component.edges]
node_labels = [component.nodes[node]['label'] for node in component.nodes]
labels = edge_labels + node_labels
self.features.loc[self.features['label'].isin(labels), 'n_branches'] = n_branches
self.features.loc[self.features['label'].isin(labels), 'n_endpoints'] = n_endpoints
self.features.loc[self.features['label'].isin(labels), 'n_branch_points'] = n_branchpoints
self.features.loc[self.features['label'].isin(labels), 'n_cycle_basis'] = n_cycle_basis
self.features.loc[self.features['label'].isin(labels), 'n_possible_undirected_cycles'] = n_possible_undirected_cycles
self.features.loc[self.features['label'].isin(labels), 'n_nodes'] = n_branchpoints + n_endpoints
def analyze_skeleton_comprehensive(
labels_input: Labels,
neighborhood: str = 'n8',
viewer: 'napari.Viewer' = None) -> Labels:
"""
Run a complete Skeleton analysis using napari-toska
Parameters:
-----------
labels_data:
"""
from napari_skimage_regionprops import TableWidget
Skeleton = ToskaSkeleton(labels_data=labels_input.data,
neighborhood=neighborhood)
Skeleton.analyze()
Skeleton.name = f'Skeleton of {labels_input.name}'
if viewer is not None:
table_widget = TableWidget(viewer=viewer, layer=Skeleton)
viewer.window.add_dock_widget(table_widget)
return Skeleton