import numpy as np
_2D_NEIGHBORHOODS = ["n4", "n8"]
_3D_NEIGHBORHOODS = ["n6", "n18", "n26"]
[docs]
def generate_labeled_skeletonization(label_image:'napari.types.LabelsData')->'napari.types.LabelsData':
'''
Skeletonize a label image and relabels the skeleton to match input labels.
The used skeletonization algorithm is the scikit-image implementation of Lee, 94.
Parameters:
-----------
label_image: napari.types.LabelsData
Input napari version of numpy.ndarray containing integer labels of an instance segmentation.
Returns:
--------
labeled_skeletons: napari.types.LabelsData
A skeleton image multiplied by the respective label of the object of origin in the input label image.
'''
from skimage.morphology import skeletonize
binary_skeleton = skeletonize(label_image.astype(bool)).astype(int)
binary_skeleton = binary_skeleton / np.amax(binary_skeleton)
labeled_skeletons = binary_skeleton * label_image
return labeled_skeletons.astype(int)
[docs]
def parse_all_skeletons(skeleton_image: "napari.types.LabelsData",
neighborhood: str = "n4") -> "napari.types.LabelsData":
from skimage import measure
properties = measure.regionprops(skeleton_image)
parsed_skeletons_assembly = np.zeros_like(skeleton_image)
for prop in properties:
sub_image = prop["image"]
sub_bbox = prop["bbox"]
sub_label = prop["label"]
# pad subimages to avoid kernel failure on edges
padded_sub_skeleton = np.pad(sub_image, 1)
parsed_sub_skeleton = parse_single_skeleton(padded_sub_skeleton * sub_label,
sub_label,
neighborhood)
# remove padding for 2D or 3D image and add to assembly
if len(skeleton_image.shape) == 2:
unpadded_image = parsed_sub_skeleton[1: -1, 1: -1]
y0, x0, y1, x1 = sub_bbox
parsed_skeletons_assembly[y0: y1, x0: x1] = unpadded_image
elif len(skeleton_image.shape) == 3:
unpadded_image = parsed_sub_skeleton[1: -1, 1: -1, 1: -1]
z0, y0, x0, z1, y1, x1 = sub_bbox
parsed_skeletons_assembly[z0: z1, y0: y1, x0: x1] = unpadded_image
return parsed_skeletons_assembly
[docs]
def parse_single_skeleton(skel: "napari.types.LabelsData",
label: int,
neighborhood: str = "n4") -> "napari.types.LabelsData":
"""
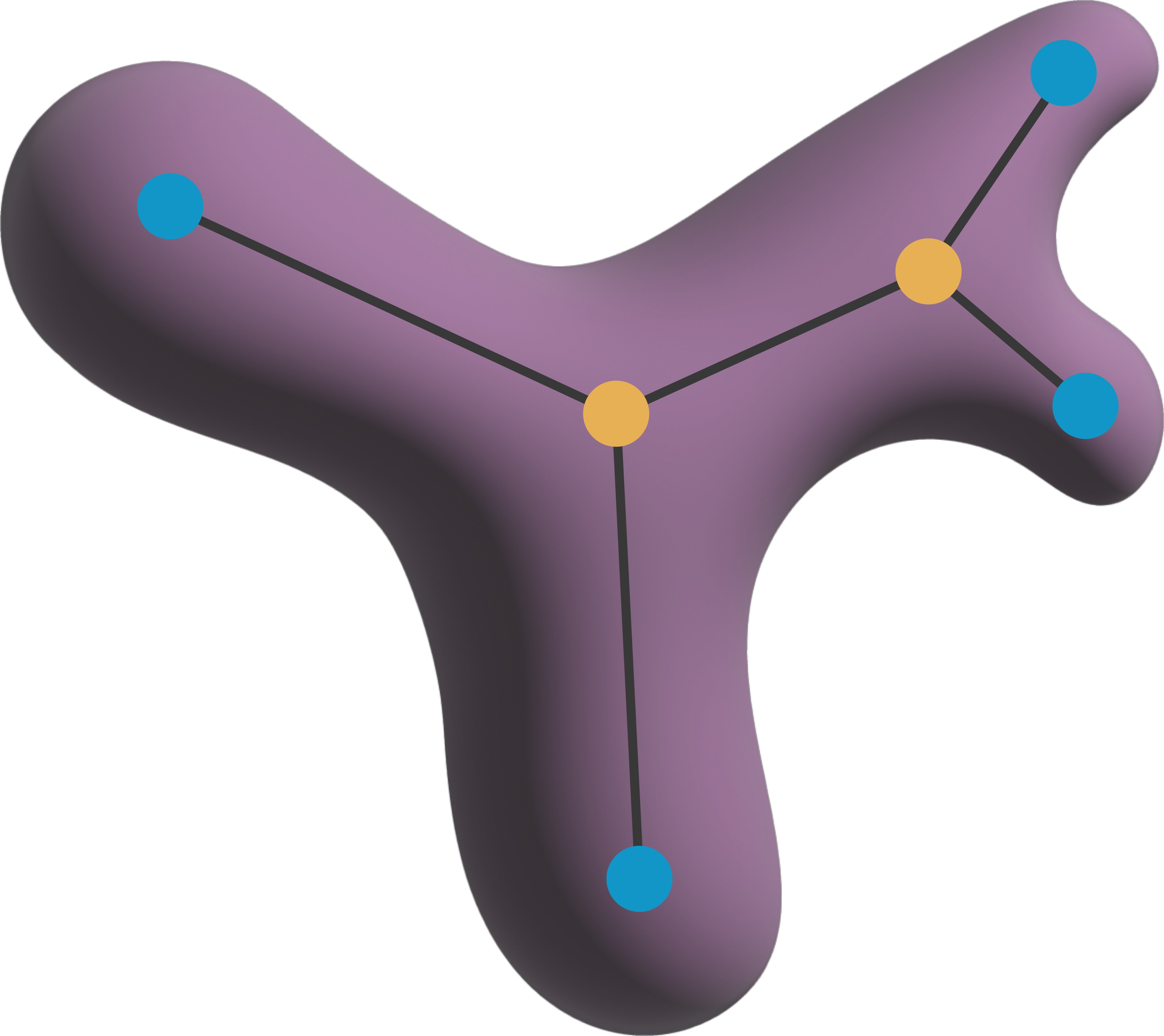
Label the skeleton of a 2D or 3D object with 4-, 6-, 8-, 18-, or 26-connectivity.
Parameters
----------
skel : napari.types.LabelsData
A skeletonized image
label : int
The label of the object whose skeleton is to be labeled.
neighborhood : str
The neighborhood connectivity of the skeleton. Can be "n4", "n6", "n8", "n18", or "n26".
Returns
-------
skeleton_labels : napari.types.LabelsData
A labeled image with the same shape as `skel` where each pixel is
labeled as either an end point (1), a branch point (2), or a
skeleton pixel (3).
"""
from ._backend_toska_functions import (n4_parse_skel_2d,
n8_parse_skel_2d,
n6_parse_skel_3d,
n18_parse_skel_3d,
n26_parse_skel_3d)
# retrieve chosen skeleton from image
skel = skel == label
if len(skel.shape) == 2:
x_dir = 1
y_dir = 0
if neighborhood == "n4":
_, e_pts, b_pts, brnch, _, _ = n4_parse_skel_2d(skel, y_dir, x_dir)
elif neighborhood == "n8":
_, e_pts, b_pts, brnch, _, _ = n8_parse_skel_2d(skel, y_dir, x_dir)
elif len(skel.shape) == 3:
x_dir = 2
y_dir = 1
z_dir = 0
if neighborhood == "n6":
_, e_pts, b_pts, brnch, _, _ = n6_parse_skel_3d(skel, z_dir, y_dir, x_dir)
elif neighborhood == "n18":
_, e_pts, b_pts, brnch, _, _ = n18_parse_skel_3d(skel, z_dir, y_dir, x_dir)
elif neighborhood == "n26":
_, e_pts, b_pts, brnch, _, _ = n26_parse_skel_3d(skel, z_dir, y_dir, x_dir)
skeleton_labels = np.zeros_like(skel, dtype=int)
skeleton_labels[brnch > 0] = 2
if len(skel.shape) == 2:
for pt in b_pts:
skeleton_labels[pt[0], pt[1]] = 3
for pt in e_pts:
skeleton_labels[pt[0], pt[1]] = 1
elif len(skel.shape) == 3:
for pt in b_pts:
skeleton_labels[pt[0], pt[1], pt[2]] = 3
for pt in e_pts:
skeleton_labels[pt[0], pt[1], pt[2]] = 1
return skeleton_labels
[docs]
def label_branches(parsed_skeletons: "napari.types.LabelsData",
labelled_skeletons: "napari.types.LabelsData",
neighborhood: str
) -> "napari.types.LabelsData":
"""
Label the branches of a skeleton image.
The branch labels start with 1 and increase for each branch.
Parameters
----------
parsed_skeletons : napari.types.LabelsData
A skeleton image where each pixel is labelled according to the
point type which can be either a terminal point (1), a branching
point (3), or a chain point (2).
labelled_skeletons : napari.types.LabelsData
A skeleton image with each skeleton carrying a unique label.
Returns
-------
branch_label_image : napari.types.LabelsData
A skeleton image where each branch is labelled with a skeleton-wise
unique label. The branch labels of each skeleton start with 1 and
increase for each branch up to the number of branches in the
skeleton.
"""
import numpy as np
from scipy import ndimage
from ._utils import get_neighborhood
structure = get_neighborhood(neighborhood)
branch_image = parsed_skeletons == 2
skeletons_ids = np.arange(1, np.max(labelled_skeletons) + 1)
branch_label_image = np.zeros_like(labelled_skeletons, dtype=int)
for i in skeletons_ids:
sub_skeleton_branches = branch_image * (labelled_skeletons == i)
branch_label_image = ndimage.label(
sub_skeleton_branches,
structure=structure)[0] + branch_label_image
return branch_label_image
[docs]
def generate_labeled_outline(labels: 'napari.types.LabelsData'):
"""
Generate a labeled outline of a label image.
Parameters
----------
labels : napari.types.LabelsData
A label image.
Returns
-------
outline : napari.types.LabelsData
A labeled outline of the label image.
"""
from skimage import segmentation, morphology
outline = segmentation.find_boundaries(
labels > 0,
connectivity=labels.ndim, mode='inner')
outline = morphology.skeletonize(outline)
return outline * labels
[docs]
def generate_local_thickness_skeleton(
outline: 'napari.types.LabelsData',
labeled_skeleton: 'napari.types.LabelsData'
) -> 'napari.types.ImageData':
"""
Generate a local thickness image of a skeleton.
Parameters
----------
outline : napari.types.LabelsData
A labeled outline of a label image.
labeled_skeleton : napari.types.LabelsData
A labeled skeleton image.
Returns
-------
thickness_image : napari.types.ImageData
A local thickness image of the skeleton.
"""
from scipy.spatial.distance import cdist
skeleton_coordinates = np.stack(np.where(labeled_skeleton > 0)).T
boundary_coordinates = np.stack(np.where(outline > 0)).T
distances_images = np.zeros_like(labeled_skeleton, dtype=np.float32)
for point in skeleton_coordinates:
distance = min(cdist([point], boundary_coordinates).squeeze())
distances_images[point[0], point[1]] = distance
return distances_images
[docs]
def reconstruct_from_local_thickness(
thickness_image: 'napari.types.ImageData'
) -> 'napari.types.LabelsData':
"""
Reconstruct a label image from a local thickness image.
Parameters
----------
thickness_image : napari.types.ImageData
A local thickness image of a skeleton.
Returns
-------
reconstruction : napari.types.LabelsData
A label image reconstructed from the local thickness image.
"""
from skimage import draw
reconstruction = np.zeros_like(thickness_image, dtype=int)
coordinates_skeleton = np.stack(np.where(thickness_image > 0)).T
for pt in coordinates_skeleton:
thickness = thickness_image[tuple(pt)]
if thickness_image.ndim == 3:
sphere = np.where(draw.ellipsoid(thickness, thickness, thickness) > 0)
# add offset to coordinates
sphere = np.stack([sphere[0] + pt[0],
sphere[1] + pt[1],
sphere[2] + pt[2]], axis=1)
# overwrite values at coordinates
reconstruction[sphere[:, 0],
sphere[:, 1],
sphere[:, 2]] = 1
elif thickness_image.ndim == 2:
disk = draw.disk(pt, thickness, shape=reconstruction.shape)
reconstruction[disk] = 1
return reconstruction