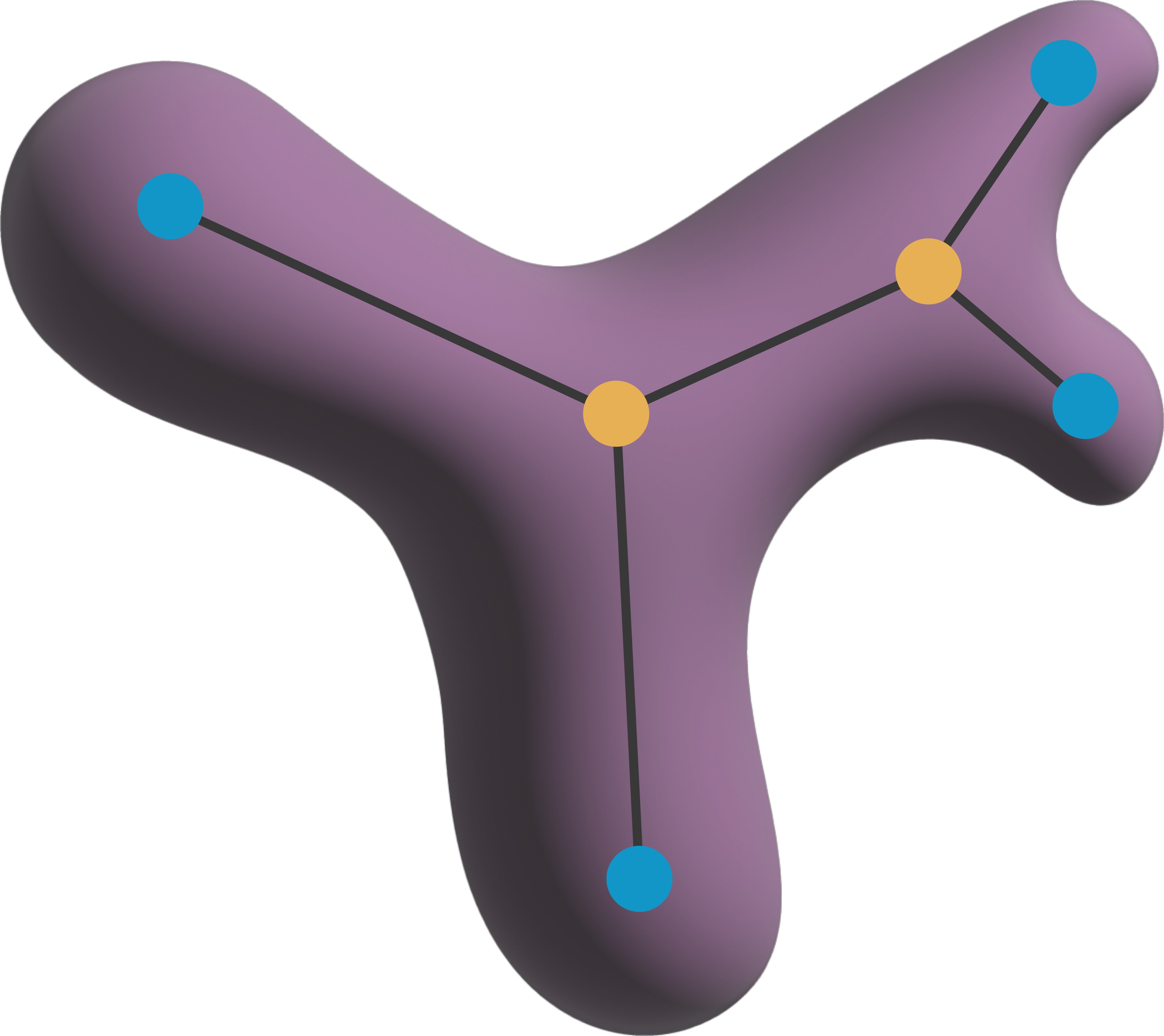
import pandas as pd
[docs]
def analyze_skeletons(
labeled_skeletons: "napari.types.LabelsData",
parsed_skeletons: "napari.types.LabelsData",
neighborhood: str = "n8",
viewer: "napari.Viewer" = None
) -> pd.DataFrame:
"""
Analyze a skeleton image and return a pandas dataframe.
This function runs the `analyze_single_skeleton` function for every
skeleton in the image and returns a pandas dataframe containing the
measurements.
Parameters
----------
labeled_skeletons : "napari.types.LabelsData"
A labeled image of skeletons.
parsed_skeletons : "napari.types.LabelsData"
A parsed labeled image of skeletons.
neighborhood : str, optional
The neighborhood used for the skeletonization, by default "n8".
For 2D images, use "n4" or "n8".
For 3D images, use "n6", "n18" or "n26".
Returns
-------
df_all : pd.DataFrame
A pandas dataframe containing the measurements.
"""
import numpy as np
for label in np.unique(labeled_skeletons)[1:]:
parsed_skeletons_single = parsed_skeletons * (labeled_skeletons == label)
df = analyze_single_skeleton(
parsed_skeletons_single, neighborhood=neighborhood)
df["skeleton_id"] = label
if label == 1:
df_all = df
else:
df_all = pd.concat([df_all, df], axis=0)
# move skeleton id to first column by its name
col = df_all.pop('skeleton_id') # Remove column 'B' and store it in col
df_all.insert(0, 'skeleton_id', col)
# add label column
df_all["label"] = df_all["skeleton_id"].values
if viewer is not None:
from napari_workflows._workflow import _get_layer_from_data
from napari_skimage_regionprops import add_table
skeleton_layer = _get_layer_from_data(viewer, labeled_skeletons)
skeleton_layer.features = df_all
add_table(skeleton_layer, viewer)
return df_all
[docs]
def analyze_single_skeleton(
parsed_skeleton: "napari.types.LabelsData",
neighborhood: str = "n8"
) -> pd.DataFrame:
"""
Analyze a single skeleton and return a pandas dataframe.
This function calculates the following measurements for a single skeleton:
- number of end points
- number of branch points
- number of nodes
- number of branches
- spine length (in network), number of edges in spine
- spine length (in image), number of pixels in spine
- number of cycle basis
- number of possible undirected cycles
Parameters
----------
parsed_skeleton : "napari.types.LabelsData"
A parsed labeled image of a skeleton.
neighborhood : str, optional
The neighborhood used for the skeletonization, by default "n8".
For 2D images, use "n4" or "n8".
For 3D images, use "n6", "n18" or "n26".
Returns
-------
df : pd.DataFrame
A pandas dataframe containing the measurements.
"""
import networkx as nx
import numpy as np
from ._network_analysis import (
create_adjacency_matrix,
convert_adjacency_matrix_to_graph,
create_spine_image)
from ._labeled_skeletonization import (
label_branches
)
from ._backend_toska_functions import skeleton_spine_search
# create an adjacency matrix for the skeleton
adjacency_matrix = create_adjacency_matrix(parsed_skeleton,
neighborhood=neighborhood)
graph = convert_adjacency_matrix_to_graph(adjacency_matrix)
# number of end points
n_endpoints = sum(adjacency_matrix.sum(axis=1) == 1)
# number of branch points
n_branch_points = sum(adjacency_matrix.sum(axis=1) > 1)
# number of nodes
n_nodes = n_endpoints + n_branch_points
# number of branches
n_branches = adjacency_matrix.shape[1]
# # neighboring end points need to be handled separately
# if n_branches == 0:
# spine_length = 0
# image_spine_length = 0
# else:
# # spine length (in network), number of edges in spine
# _, spine_paths_length = skeleton_spine_search(
# adjacency_matrix, graph)
# spine_length = np.nansum(spine_paths_length)
# # spine length (in image), number of pixels in spine
# branch_labels = label_branches(parsed_skeleton, parsed_skeleton > 0,
# neighborhood=neighborhood)
# spine_image = create_spine_image(adjacency_matrix, branch_labels)
# image_spine_length = calculate_spine_length(spine_image)
# cycle basis
directed_graph = graph.to_directed()
possible_directed_cycles = list(nx.simple_cycles(directed_graph))
n_cycle_basis = len(nx.cycle_basis(graph))
n_possible_undirected_cycles = len(
[x for x in possible_directed_cycles if len(x) > 2])//2
df = pd.DataFrame(
{
"n_endpoints": [n_endpoints],
"n_branch_points": [n_branch_points],
"n_nodes": [n_nodes],
"n_branches": [n_branches],
# "spine_length_network": [spine_length],
# "spine_length_image": [image_spine_length],
"n_cycle_basis": [n_cycle_basis],
"n_possible_undirected_cycles": [n_possible_undirected_cycles]
}
)
return df
[docs]
def analyze_single_skeleton_network(
parsed_skeleton_single: "napari.types.LabelsData",
neighborhood: str = "n8"
) -> pd.DataFrame:
"""
Analyze a single skeleton and return a pandas dataframe.
This function categorizes ever element of the network
representation of a skeleton as either a node or an edge
and potentially its weight.
Parameters
----------
parsed_skeleton_single : "napari.types.LabelsData"
A parsed labeled image of a skeleton.
neighborhood : str, optional
The neighborhood used for the skeletonization, by default "n8".
For 2D images, use "n4" or "n8".
For 3D images, use "n6", "n18" or "n26".
Returns
-------
features : pd.DataFrame
A pandas dataframe containing the measurements.
"""
import networkx as nx
import numpy as np
from skimage import measure
import pandas as pd
from ._network_analysis import (
create_adjacency_matrix,
convert_adjacency_matrix_to_graph)
from ._backend_toska_functions import skeleton_spine_search
# create an adjacency matrix for the skeleton
adjacency_matrix = create_adjacency_matrix(parsed_skeleton_single,
neighborhood=neighborhood)
graph = convert_adjacency_matrix_to_graph(adjacency_matrix)
# get edge and node labels
node_labels = np.arange(1, adjacency_matrix.shape[0]+1)
edge_labels = np.arange(1, adjacency_matrix.shape[1]+1)
# component type
component_type = ['node'] * adjacency_matrix.shape[0] +\
['edge'] * adjacency_matrix.shape[1]
# Assemble the table
features = pd.DataFrame(
{
"label": np.arange(1, np.amax(node_labels) + np.amax(edge_labels) +1),
"component_type": component_type
}
)
# Measurement: Node degree
node_degrees = adjacency_matrix.sum(axis=1)
features.loc[features["component_type"] == "node", "degree"] = node_degrees
# add all edge weights to dataframe
edge_weights = nx.get_edge_attributes(graph, "weight")
features.loc[features["component_type"] == "edge", "weight"] = list(edge_weights.values())
features.loc[features["component_type"] == "edge", "node_1"] = np.asarray(graph.edges)[:, 0]
features.loc[features["component_type"] == "edge", "node_2"] = np.asarray(graph.edges)[:, 1]
features.loc[features["component_type"] == "node", "node_labels"] = list(graph.nodes)
return features
[docs]
def calculate_branch_lengths(
branch_label_image: "napari.types.LabelsData",
viewer: "napari.Viewer" = None
) -> pd.DataFrame:
"""
Calculate the branch length for each branch in a branch image.
This function calculates the branch length for each branch in a
branch image. The branch length is calculated as the number of
pixels in the branch and takes into account the adjacency
relationship between subsequent pixels/voxels in a branch.
Parameters
----------
branch_label_image : "napari.types.LabelsData"
A labeled image of an individual skeleton's branches.
Returns
-------
df : pd.DataFrame
A pandas dataframe containing the branch length for each branch.
"""
import numpy as np
from skimage import measure
from copy import deepcopy
unique_branches = np.unique(branch_label_image)[1:]
df = pd.DataFrame(
{
"label": unique_branches,
}
)
if len(branch_label_image.shape) == 2:
# loop over all branches
for branch in unique_branches:
length = 0
masked_branch = branch_label_image * (branch_label_image == branch)
labeled_branch = measure.label(masked_branch, connectivity=1)
unique_branch_segments = np.unique(labeled_branch)[1:]
# loop over all segments of the branch
for label in unique_branch_segments:
segment_size = np.sum(unique_branch_segments == label)
if segment_size == 1:
length += np.sqrt(2)
else:
length += segment_size
df.loc[df["label"] == branch, "branch_length"] = length
elif len(branch_label_image.shape) == 3:
for branch in unique_branches:
length = 0
masked_branch = branch_label_image * (branch_label_image == branch)
n6_labeled_branch = measure.label(masked_branch, connectivity=1)
n6_labeled_branch_copy = deepcopy(n6_labeled_branch)
n6_unique_branch_segments = np.unique(n6_labeled_branch)[1:]
# loop over all segments of the branch
for segment in n6_unique_branch_segments:
segment_size = np.sum(n6_labeled_branch == segment)
if segment_size == 1:
n6_labeled_branch_copy[n6_labeled_branch == segment] = 0
face_sharing_contributions = np.sum(n6_labeled_branch_copy > 0)
length += face_sharing_contributions
n18_labeled_branch = measure.label(masked_branch, connectivity=2)
n18_unique_branch_segments = np.unique(n18_labeled_branch)[1:]
# loop over all segments of the branch
for segment in n18_unique_branch_segments:
segment_size = np.sum(n18_labeled_branch == segment)
if segment_size == 1:
length += np.sqrt(3)
n18_labeled_branch[n18_labeled_branch == segment] = 0
n18_remainder_objects = n18_labeled_branch - n6_labeled_branch_copy
edge_sharing_contributions = np.sum(n18_remainder_objects > 0)
length += edge_sharing_contributions * np.sqrt(2)
df.loc[df["label"] == branch, "branch_length"] = length
if viewer is not None:
from napari_workflows._workflow import _get_layer_from_data
from napari_skimage_regionprops import add_table
branch_layer = _get_layer_from_data(viewer, branch_label_image)
branch_layer.features = df
add_table(branch_layer, viewer)
return df
def calculate_spine_length(
spine_image: "napari.types.LabelsData",
) -> float:
"""
Iterate over every present label in the spine image and calculate
the spine length for each label.
Parameters
----------
spine_image : "napari.types.LabelsData"
A labeled image of a skeleton's spine.
Returns
-------
total_length : float
The total length of the spine.
"""
segment_lengths = calculate_branch_lengths(spine_image)
total_length = segment_lengths["branch_length"].sum()
return total_length