Source code for napari_toska._network_analysis
import numpy as np
import networkx as nx
[docs]
def create_all_adjancency_matrices(
labeled_skeletons: "napari.types.LabelsData",
parsed_skeletons: "napari.types.LabelsData",
neighborhood: str
):
"""
Calculate all adjacency matrices for a given skeleton image.
Parameters:
-----------
labeled_skeletons: napari.types.LabelsData
A skeleton image with each skeleton carrying a unique label.
parsed_skeletons: napari.types.LabelsData
A skeleton image where each pixel is labelled according to the
point type which can be either a terminal point (1), a branching
point (3), or a chain point (2).
neighborhood: str
The neighborhood connectivity of the skeleton. Can be "n4",
"n6", "n8", "n18", or "n26".
Returns:
--------
adjacency_matrices: list
A list of adjacency matrices for each skeleton in the image.
"""
skeleton_ids = np.unique(labeled_skeletons)[1:]
adjacency_matrices = [0] * len(skeleton_ids)
skeleton_counter = 0
for skeleton_id in skeleton_ids:
# create a sub-skeleton image with only one skeleton with values
# 0 (background) and 1 (skeleton end point) etc.
sub_skeleton = parsed_skeletons * (labeled_skeletons == skeleton_id)
adjacency_matrices[skeleton_counter] = create_adjacency_matrix(sub_skeleton, neighborhood)
skeleton_counter += 1
return adjacency_matrices
[docs]
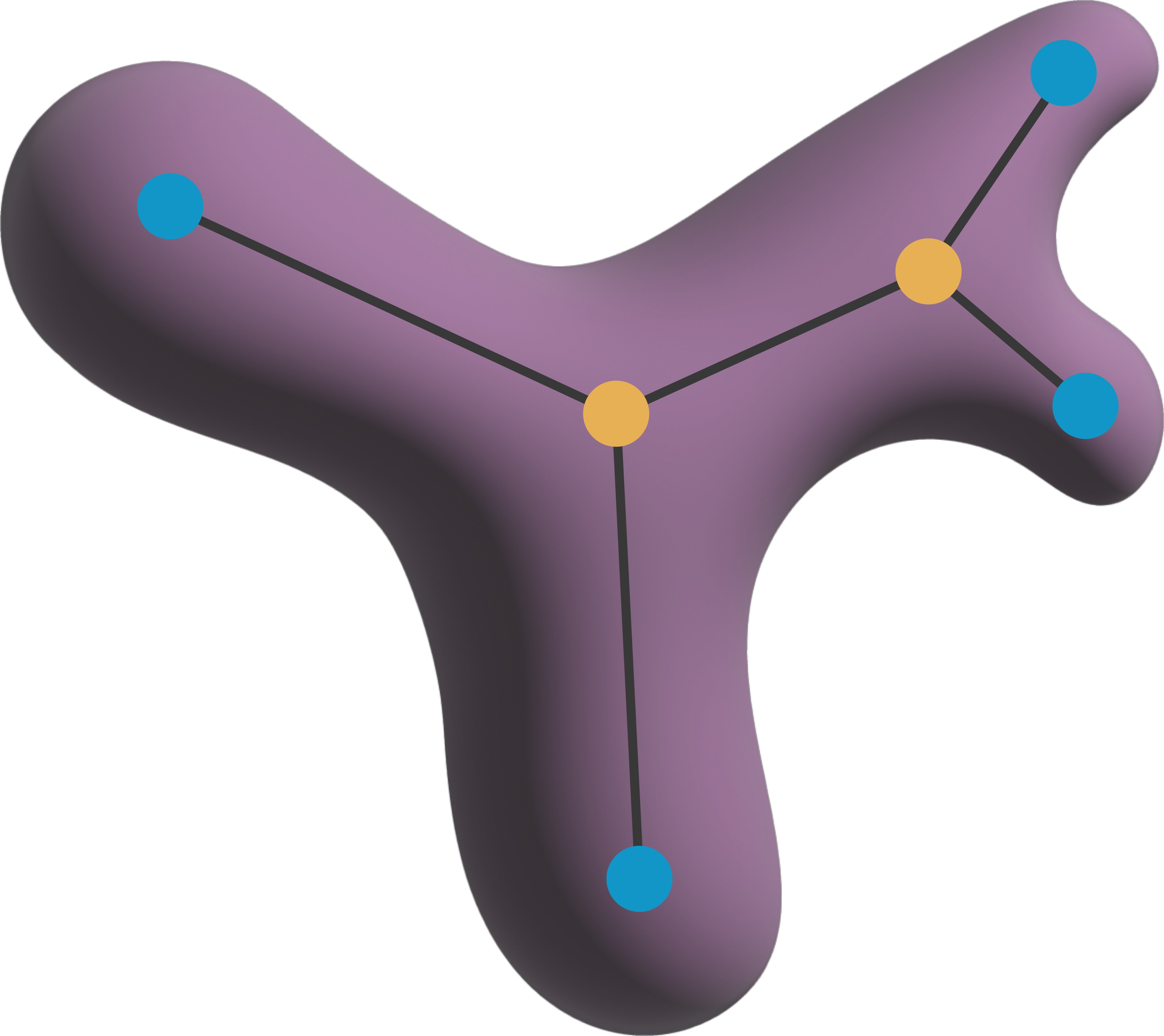
def create_adjacency_matrix(parsed_skeletons: "napari.types.LabelsData",
neighborhood: str = "n8") -> np.ndarray:
"""
Create an adjacency matrix for a given skeleton image.
Parameters:
-----------
parsed_skeletons: napari.types.LabelsData
A skeleton image where each pixel is labelled according to the
point type which can be either a terminal point (1), a branching
point (3), or a chain point (2).
neighborhood: str
The neighborhood connectivity of the skeleton. Can be "n4",
"n6", "n8", "n18", or "n26".
Returns:
--------
M: np.ndarray
An adjacency matrix for the skeleton.
"""
from scipy import ndimage
from ._backend_toska_functions import _generate_adjacency_matrix
from ._utils import get_neighborhood
structure = get_neighborhood(neighborhood)
# Retrieve branch points
branch_points = parsed_skeletons == 3
branch_points, _ = ndimage.label(
branch_points, structure=structure)
# Retrieve end points
end_points = np.asarray(np.where(parsed_skeletons == 1)).T
# Retrieve branches image
branches, _ = ndimage.label(parsed_skeletons == 2, structure=structure)
# generate adjacency matrix
M = _generate_adjacency_matrix(end_points, branch_points, branches, structure)
return M
[docs]
def convert_adjacency_matrix_to_graph(
adj_mat: np.ndarray,
weights: np.ndarray = None
) -> nx.Graph:
"""
Convert an adjacency matrix to a networkx graph.
Parameters
----------
adj_mat : numpy.ndarray
An adjacency matrix where each row represents a branching- or endpoint and
each column represents a branch. A value of 1 indicates that the
branching point or endpoint is connected to the branch.
weights : numpy.ndarray, optional
An array of weights for each branch, by default 1 (unweighted)
Returns
-------
G : networkx.Graph
A networkx graph where each node represents a branching- or endpoint and
each edge represents a branch.
"""
if weights is None:
weights = np.ones((adj_mat.shape[1]))
nodes = adj_mat
weighted_edges = []
for i in range(nodes.shape[1]):
edge = list(np.where(nodes[:, i])[0])
if len(edge) == 2:
edge.append(weights[i])
weighted_edges.append(tuple(edge))
else:
continue
G = nx.Graph()
G.add_weighted_edges_from(weighted_edges)
return G
[docs]
def create_spine_image(
adjacency_matrix: np.ndarray,
labeled_branches: "napari.types.LabelsData"
) -> "napari.types.LabelsData":
"""
Create an image where the pixels of the spine are labeled with the
branch ID of the branch they belong to.
Parameters
----------
labeled_branches : napari.types.LabelsData
A labeled image with labelled skeleton branches.
img_spine_ids : list
A list of branch labels that represent a path in a skeleton graph.
Returns
-------
img_spine : napari.types.LabelsData
A labeled image with the same shape as `labeled_branches` where
the pixels of the spine are labeled with the branch ID of the branch
they belong to.
"""
from ._backend_toska_functions import (
skeleton_spine_search,
find_spine_edges,
map_spine_edges_to_skeleton
)
# for skeletons consisting only of a single point
if (labeled_branches > 0).sum() == 1:
return labeled_branches
G = convert_adjacency_matrix_to_graph(adjacency_matrix)
spine_path_nodes, spine_path_length = skeleton_spine_search(
adjacency_matrix, G)
spine_edges = find_spine_edges(spine_path_nodes)
if len(spine_edges) == 1:
branch_labels = [1]
else:
branch_labels = np.arange(1, np.amax(labeled_branches))
spine_edges = map_spine_edges_to_skeleton(
spine_edges,
adjancency_matrix=adjacency_matrix,
branch_labels=branch_labels)
img_spine = np.zeros_like(labeled_branches)
for i in spine_edges:
img_spine[labeled_branches == i] = i
return img_spine